April 9, 2019
New electron microscopy technique limits membrane destruction
 Purdue researchers Seema Mattoo and Ranjan Sengupta have developed a new electron microscopy technique that allows them to accurately track membrane proteins without damaging the cellular environment. (Purdue University photo/Rebecca Wilcox)
Download image
Purdue researchers Seema Mattoo and Ranjan Sengupta have developed a new electron microscopy technique that allows them to accurately track membrane proteins without damaging the cellular environment. (Purdue University photo/Rebecca Wilcox)
Download image
WEST LAFAYETTE, Ind. — Membrane proteins play an important role in many biological processes. Studies suggest they’re targeted by more than 50% of all modern medicinal drugs. Unfortunately for researchers, determining their structures has been a longstanding challenge because it’s difficult to track the protein without damaging the cellular membrane using current techniques. Not anymore.
Researchers at Purdue University have created an electron microscopy technique termed “cryoAPEX” that accurately tracks membrane proteins in a well-preserved cell. The new method is a hybrid of two commonly used methods in cell biology: high-pressure freezing and chemical fixation.
“We took the best features from each technique and played around with conditions until we found a happy medium where you could stain for your protein while maintaining membrane preservation,” said Seema Mattoo, an assistant professor of biological sciences at Purdue. “We were also able to use this information to develop a 3D image of the protein in the context of the cell.”
Chemical fixation uses antibodies to detect proteins, but to do that, the cells need to be “fixed” with alcohol. Alcohol breaks down the membrane, essentially creating holes in it and allowing the signal researchers are looking for to leak out. This produces faulty results.
High-pressure freezing, on the other hand, preserves the entire cell perfectly. But this method isn’t compatible with staining techniques, so there’s no way to pinpoint the protein of interest.
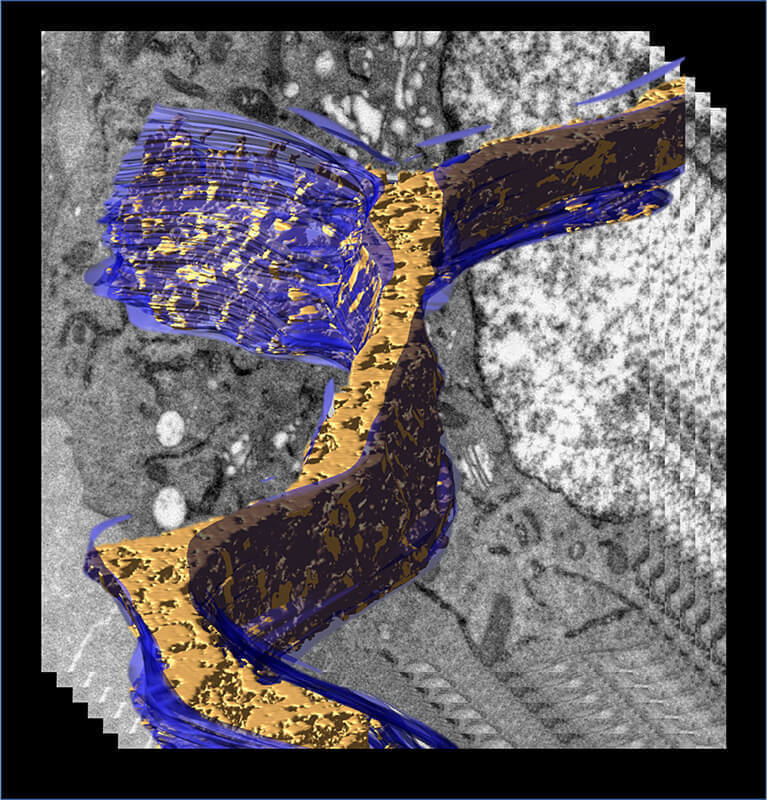 A 3D model of the endoplasmic reticulum (blue) displaying density associated with transiently expressed APEX2-tagged human Fic protein was generated from an electron tomogram of a cell processed by Cryo-APEX. (Purdue University/Seema Mattoo)
Download image
A 3D model of the endoplasmic reticulum (blue) displaying density associated with transiently expressed APEX2-tagged human Fic protein was generated from an electron tomogram of a cell processed by Cryo-APEX. (Purdue University/Seema Mattoo)
Download image
Membrane proteins play an important role in several biological processes, including relaying signals between a cell’s internal and external environments, transporting molecules and ions across the membrane, and allowing cells to identify and interact with each other. Being able to follow these proteins as they perform their duties in the cell will allow researchers to understand cell signaling mechanisms that regulate protein function.
“Virologists, for instance, will find this technique really useful because now they’ll be able to follow their viruses in the context of a particular viral protein within a cell,” said Mattoo, who is also a member of Purdue’s Center for Cancer Research and Institute of Inflammation, Immunology and Infectious Disease.
Mattoo’s research focuses on a class of enzymes called Fic proteins. The version present in humans, HYPE, is a critical regulator of whether cells under stress live or die. Her goal for developing this technique was to be able to see where HYPE goes once it enters the endoplasmic reticulum, she said.
The findings, which were published in the Journal of Cell Science, show that HYPE is strongly drawn to the lumen. This means that HYPE is tightly regulated, and if it ever does leave the endoplasmic reticulum, it would likely be under disease-specific conditions.
“As we’re trying to figure out how to manipulate HYPE to determine whether it can be used as a therapeutic, we’re going to focus a lot of our energy on the endoplasmic reticulum,” Mattoo said. “But the wider implications of these findings are predominantly in the technique. We developed it so that it can be applied to tissue culture cells and can be used by a wide range of researchers.”
This work aligns with Purdue's Giant Leaps celebration, acknowledging the university’s global advancements made in health, longevity and quality of life as part of Purdue’s 150th anniversary. This is one of the four themes of the yearlong celebration’s Ideas Festival, designed to showcase Purdue as an intellectual center solving real-world issues.
The research was supported by the National Institutes of Health, the Indiana Clinical and Translational Sciences Institute, and the Purdue Institute for Inflammation, Immunology and Infectious Disease.
Writer: Kayla Zacharias, 765-494-9318, kzachar@purdue.edu
Source: Seema Mattoo, 765-496-7293, smattoo@purdue.edu
Note to Journalists: For a copy of the paper, please contact Kayla Zacharias, Purdue News Service, kzachar@purdue.edu
ABSTRACT
CryoAPEX – an electron tomography tool for subcellular localization of membrane proteins
Ranjan Sengupta, Michael J. Poderycki, Seema Mattoo
We describe a method, termed cryoAPEX, which couples chemical fixation and high-pressure freezing of cells with peroxidase tagging (APEX) to allow precise localization of membrane proteins in the context of a well-preserved subcellular membrane architecture. Further, cryoAPEX is compatible with electron tomography. As an example, we apply cryoAPEX to obtain a high-resolution three- dimensional contextual map of the human FIC (filamentation induced by cAMP) protein, HYPE (also known as FICD). HYPE is a single- pass membrane protein that localizes to the endoplasmic reticulum (ER) lumen and regulates the unfolded protein response. Alternate cellular locations for HYPE have been suggested. CryoAPEX analysis shows that, under normal and/or resting conditions, HYPE localizes robustly within the subdomains of the ER and is not detected in the secretory pathway or other organelles. CryoAPEX is broadly applicable for assessing both lumenal and cytosol-facing membrane proteins.

