Viruses

|
Computational Studies on Viruses and Binding of Antiviral Agents
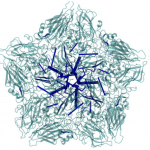
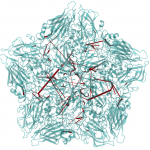
It is known that antiviral agents against human rhinovirus (HRV) inhibit uncoating of the viral capsid, a process to release genomic RNA during infection. An interest of Post is to understand how drugs enter an interior binding pocket and, once bound, how the drug alters conformational and fluctuation properties of the virus capsid to give rise to the observed antiviral activity. Starting with an observation from molecular dynamics simulations that antiviral compounds increase thermodynamic compressibility, Post postulated that the basis for antiviral activity is entropic stabilization, a proposal that later was shown experimentally to be the case. Other studies of the Post research group elucidated the dissociation process of the antiviral agent from a buried hydrophobic cavity of HRV using a perturbative, molecular dynamics simulation approach. Most recently, her group has discovered a set of very long distance correlation paths in HRV capsid that are destroyed by an antiviral compound, and thus implicated in the motions associated with uncoating. These motions and pathways suggest a previously unrecognized mechanism for uncoating.
|
 |
Viral Structural Proteins
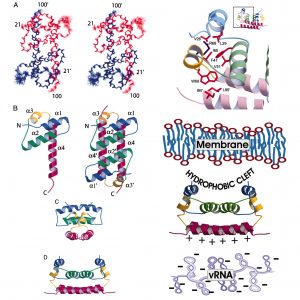
Enveloped viruses comprise a protein-nucleic acid core covered by a membrane bilayer with integral membrane receptor proteins. NMR studies by the research group of Post to define the structure and dynamics of flavivirus capsid protein, and the association of capsid and nucleocapsid from alphavirus have helped to characterize protein interactions in assembly. An antiviral compound, designed to inhibit maturation of assembled dengue virus, was discovered using computational screening of the envelope protein, followed by validation using NMR. Recent studies on the retrovirus rous sarcoma virus exploited NMR relaxation to characterize domain behavior of the gag polyprotein, which lead to the proposal for an autoinhibitory mechanism in controlling assembly of rous sarcoma virus.
|